Basics of usage
See code in GitLab.
Author: Gernot Plank gernot.plank@medunigraz.at
Overview
cd ${TUTORIALS}/02_EP_tissue/01_basic_usageThis example introduces to the basics of using the openCARP executable for simulating EP at the tissue and organ scale. Details on how to inquire input parameters are provided in the openCARP section of the manual.
In absence of pacemaker currents electrical stimulus currents must be supplied in order to depolarize tissue above the threshold to elicit a propagated action potential. The simplest way of initiating a propagating action potential is the use of a transmembrane stimulus, \(I_{\mathrm{tr}}\). According to the definition in openCARP supplying a positive \(I_{\mathrm{tr}}\) depolarizes the tissue, a negative \(I_{\mathrm{tr}}\) hyperpolarizes the tissue.
Experimental setup
To introduce definition and usage of transmembrane stimulus currents in openCARP a simple experiment using a thin monolayer preparation was defined. The following input parameters are available to steer the experiment:
--duration
Duration of simulation (ms)
--S1-strength
pick transmembrane current stimulus strength in
[uA/cm^2] = [pA/pF] considering fixed Cm (default: 20.)
--S1-dur
pick transmembrane current stimulus duration in [msModeling subthreshold conditions
Experiment exp01 (subthreshold depolarizing stimulus)
We start with a longer stimulus lasting for 15 ms and gradually increase the stimulus strength until we achieve capture. This is equivalent to measuring one data point of a strength-duration curve to determine chronaxie and rheobase of our preparation.
./run.py --duration 20 --S1-strength 20. --S1-dur 15 --visualizeThis weak current slowly depolarizes the membrane, but does not reach threshold.
After the end of stimulation, the membrane returns to its resting state, as shown in fig-tutorial-subthreshold-depol-stim.
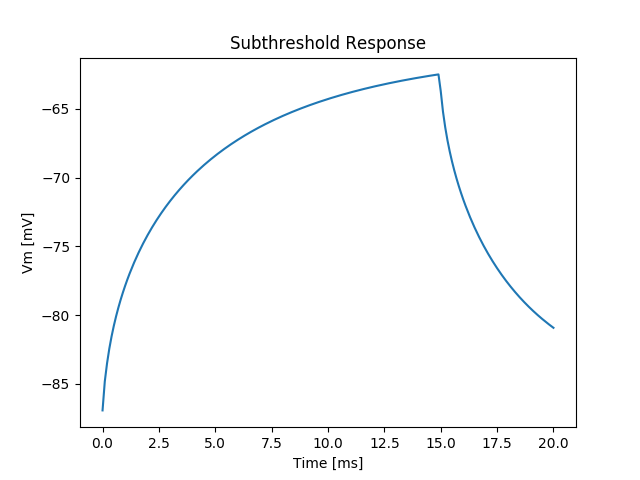
Inspect the variation in membrane depolarization as a function of distance to the stimulus location by varying the vertex index in meshalyzer.
To understand how the stimulus definition is passed on to the simulator we inspect the command line generated by the script. For this sake run
./run.py --duration 20 --S1-strength 20. --S1-dur 15 --dryThe stimulus section of the command line is then
-num_stim 1 \ # use one stimulus electrode
-stim[0].name S1 \ # pulse and type definition
-stim[0].crct.type 0 -stim[0].pulse.strength 20.0 -stim[0].ptcl.duration 15.0 \ # electrode definition (default=block)
-stim[0].elec.p0[0] -5050.0 -stim[0].elec.p1[0] -4950.0 -stim[0].elec.p0[1] -550.0 -stim[0].elec.p1[1] 550.0 -stim[0].elec.p0[2] -150.0 -stim[0].elec.p1[2] 150.0Experiment exp02 (subthreshold hyperpolarizing stimulus)
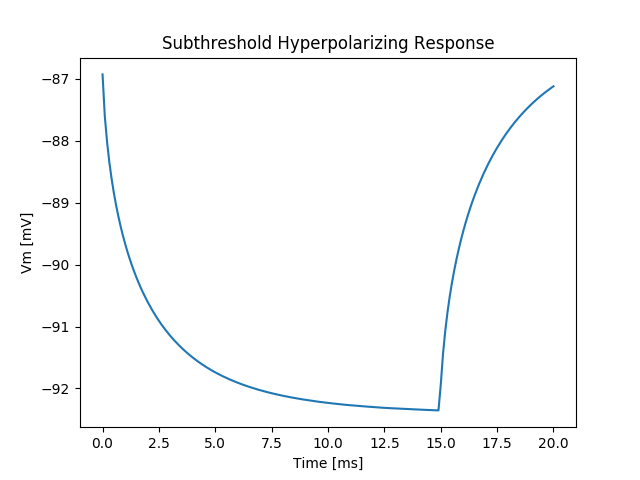
We repeat exp01 with a negative transmembrane stimulus current.
./run.py --duration 20 --S1-strength -20 --S1-dur 15 --visualizeAs the membrane behaves essentially only passively in the subthreshold regime
we observe the same response symmetric to the resting potential, see fig-tutorial-subthreshold-hyper-stim
Initiating action potential propagation
We repeat exp01 with a stronger transmembrane stimulus current of shorter duration.
Peak sodium current is known to be around \(250 \mu A/cm^2\) and last for less than a millisecond.
We chose a strength of \(250 \mu A/cm^2\) of 1 millisecond duration to achieve suprathreshold depolarization.
Keep in mind that suprathreshold depolarization at the stimulus site does not automatically lead to a propagated response.
The local membrane response at the stimulus location is shown in fig-tutorial-suprathreshold-stim.
./run.py --duration 20 --S1-strength 250. --S1-dur 2 --visualize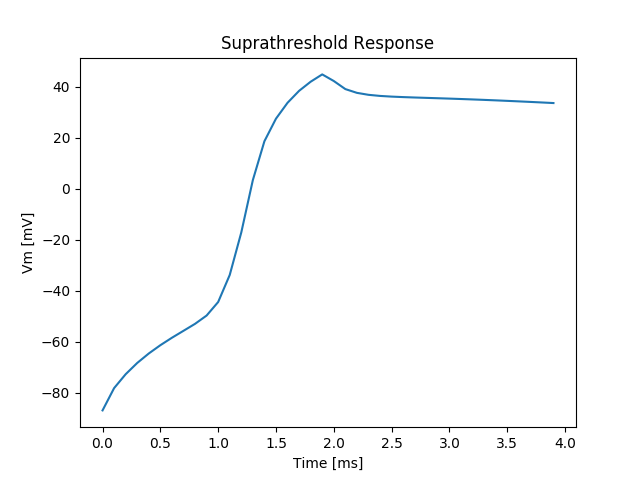
Recent questions tagged experiments, examples, stimulus, tissue
There are tagged with experiments, examples, stimulus, tissue.
Here we display the 5 most recent questions. You can click on each tag to show all questions for this tag.
You can also ask a new question.
