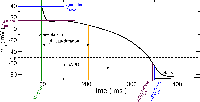openCARP examples
These examples are intended to transfer basic user know-how regarding most openCARP features in an efficient way. The scripts are designed as mini-experiments, which can also serve as basic building blocks for more complex experiments. This example can be a good starting point to base your own experiment on.
There is a number of examples dedicated to teaching openCARP fundamental know-how for those who are interested in building more complex experiments from scratch themselves or in extending pre-existing experiments. All executable examples are coded up in carputils to facilitate an easy execution of all experiments without significant additional effort and complex command line interactions.
Intended use
Most examples can be run by simply copying the command from the corresponding example web page. It is recommended to inspect the generated command lines to understand what the simulation looks like in the plain command line by adding the option --dry to the run script command line. You can download the examples from our repository.
Electrophysiology in single cell
The following examples illustrate how single cell modeling is performed using the tool bench. Additionally you learn how to integrate a single cell model from CellML into our library limpet using the math language EasyML

Basic single cell EP
This example introduces the basic steps of running EP simulations in an isolated myocytes

APD restitution
Action potenital duration (APD) restitution example in single cell. As pacing frequency is increased, APD shortens to maintain a one to one stimulus to responses
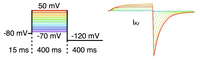
Voltage clamp
This example explains the basic usage of bench for performing voltage clamp experiments.
EasyML to C code
This tutorial explains the basics of using the code generation tool limpet_fe.py for generating ODE solver code for models of cellular dynamics. It also covers the creation of new models as a dynamic library.
EasyML basics
More information about the cell model math format EasyML
Electromechanical coupling
Couple an electrophysiological cell model to a tension model
Import CellML
Here you learn how to import CellML data into openCARP and what problems might occur during this process
Convert EasyML (.model) to CellML
Learn how to convert your .model file (EasyML) to CellML
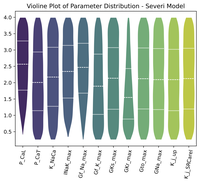
Parallel Single Cell Simulations
See how to execute single-cell simulations in parallel and build populations of models.
Electrophysiology tissue
These examples inform about basic steps in developing tissue simulations based on the monodomain and bidomain models
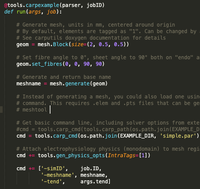
Simple example to base your own experiment on
This is not a real example but more of a template to base your own experiment on. It covers mesh generation, monodomain simulation and local activation time extraction during postprocessing.
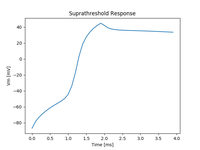
Basic tissue EP
This example introduces to the basics of using the openCARP executable for simulating EP at the tissue and organ scale
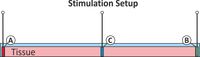
Extracellular stimulation
In this example you learn how to stimulate a tissue from the extracellular space
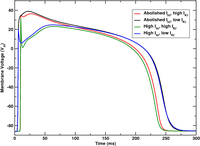
APD adjustment
This example demonstrates how to adjust ionic model parameters to generate a specific action potential duration in your simulations
Tuning conduction velocity
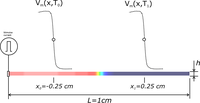
This tutorial introduces the background for the relationship between conduction velocity and tissue conductivity

Init tissue from cell
This tutorial demonstrates how to initialize a cardiac tissue with state variables obtained from a single-cell stimulation
Adjust parameters
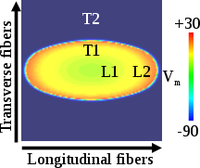
This tutorial demonstrates how to adjust parameters in tissue simulations to match experimental data for conduction velocity, APD, and wavelength
CV restitution
This example demonstrates how to compute conduction velocity restitution in cardiac tissue
Niederer et al. benchmark
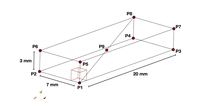
This example replicates the Niederer et al. benchmark and illustrates effects of some numerical settings including temporal and spatial discretization
ERP restitution
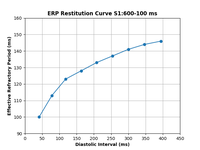
This tutorial shows how to calculate the effective refractory period (ERP) for a given combination of ionic conductances in a tissue slab using an S1S2 pacing protocol. An ERP restitution curve can also be plotted, if desired by the user.
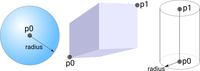
Tissue regions
Regions are used to manage the assignment of heterogeneous tissue properties. This tutorial explains the different approches of how regions can be defined.
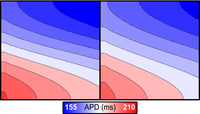
Region vs. gradient heterogeneities
This tutorial introduces the concepts of region-based and gradient-based heterogeneities for assigning spatially varying properties
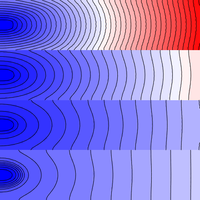
Regions & conductivities
This tutorial details how to assign different conductivities to different parts of a simulated tissue slice using region-wise tagging
EP heterogeneity
This example details how to assign different single cell dynamics to different parts of a simulated tissue slice using region-wise tagging
Region reunification
This example details how to output the values of state variables over time during a simulation
Smooth gradient
This example details how to assign a gradient of single cell properties using the adjustments interface
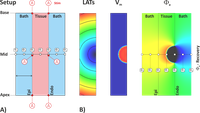
Extracellular potentials and ECGs
This tutorial explains the background of computing extracellular potentials and ECG using different techniques

Periodic boundary conditions
Periodic boundary conditions connect the left edge of a sheet to the right, or the top to the bottom
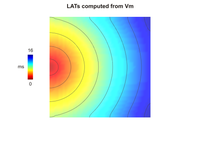
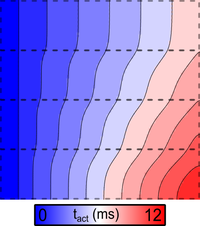
Local activation time
This example demonstrates how to compute local activation times (LATs) and action potential durations (APDs) of cells in a cardiac tissue
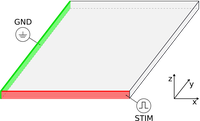
Boundary conditions
Computing Laplace-Dirichlet maps provide an elegant tool for describing the distance between defined boundaries
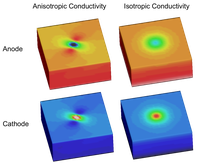
Anisotropy effects
Unequal anisotropy ratios can be responsible for the formation of unexpectedly complex polarization patterns
Parameter sweeps
This example shows how to use polling files to sweep parameters
Reentry induction
This example shows the influence of different induction protocol on reentry initiation and maintenance
Eikonal
These examples inform about basic steps in developing tissue simulations based on the eikonal, reaction-eikonal and DREAM models
Eikonal model
This example demonstrates how to set up and use the eikonal model
Reaction-Eikonal Model
This example shows how to run reaction-eikonal simulations with (RE+) and without diffusion current (RE-)
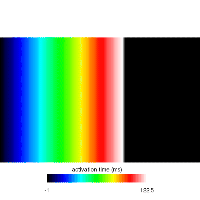
Diffusion Reaction Eikonal Alternant Model - Pacing
This example demonstrates and explores pacing behavior while using the DREAM.
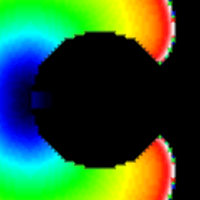
Diffusion Reaction Eikonal Alternant Model - Reentry
This example demonstrates how to set up and use the DREAM to simulate reentries
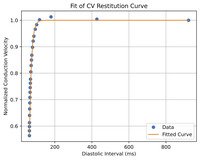
Fitting CV restitution
This example shows how to find the parameters for CV restitution to use DREAM with other ionic models
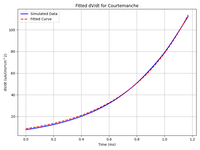
Fitting foot current parameters
This example shows how to find the parameters for Ifoot to run reaction-eikonal with other ionic models
Visualization
Learn how to use the visualization tools LimpetGUI for single cell results as well as Meshalyzer and ParaView for tissue results
limpetGUI
limpetGUI is a simple tool for visualizing traces output generated by bench
Meshalyzer
meshalyzer is a 4D visualization tool for openCARP allowing interactions to investigate data
Meshalyzer II
A few advanced features of meshalyzer are explained here to make life simpler
Paraview visualization
This is an example to visualize the time series of a simple tissue geometry in ParaView.
Pre- and post-processing
Learn how to use the meshing tools and how to postprocess igb files