Onboarding tutorials in openCARP JupyterLab
You have three options to run the onboarding tutorials in openCARP:
- Local openCARP JupyterLab using Docker on your own computer
- Our openCARP JupyterHub server
- mybinder running openCARP JupyterLab
All of them have advantages and disadvantages.
| Platform | Advantage | Disadvantage |
|---|---|---|
| Docker | Using your own resources (likely more than the other options); full control | Needing to set up docker and run container |
| JupyterHub | We take care that everything works | You need a login for openCARP GitLab; slower due to limited resources |
| mybinder | We take care that everything works | All progress is lost after some time without interaction or reload of the page; limited resources; quite long start-up time |
You will see a link to the three options in each of the cards below. If you don't see a link to the local docker version, you don't have it running. Follow the instructions, if you want to run openCARP JupyterLab in a local Docker installation. After the openCARP Docker is running, you should reload this page. If you try the local docker version on a Mac, please don't use Safari, as you wouldn't get a connection to localhost. Firefox or Chrome should work.
If you use the JupyterHub server, you have to sign-in to openCARP GitLab. Please follow the link "Sign in with openCARP Gitlab" and than use the link ->"Helmholtz AI / AAI"<-. Please search for you home institution (either in english, your language or as abbreviation) and login to your home institution. Follow the instructions on your home institutions authentication webpage. If you don't find your home institution, please search for ORCID (if you have), or GitHub (not recommended, but possible).
Single Cell Tutorials
Aim of the single cell tutorials is to make you familiar with the command line interface of the single cell electrophysiology model program bench to develop single cell experiments. You also will learn basics about carputils. We strongy suggest to go through them in sequence.
Overview page of onboarding tutorials
If you want to start from the base and see this overview directly in openCARP JupyterLab.
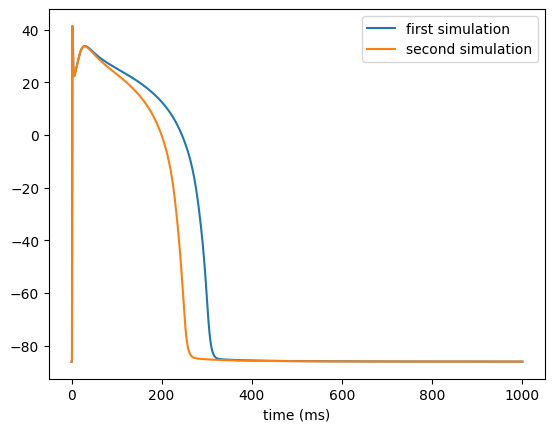
Basic functionality I
The first tutorial should familiarize you with the basic functionality of the ionic model program `bench`.
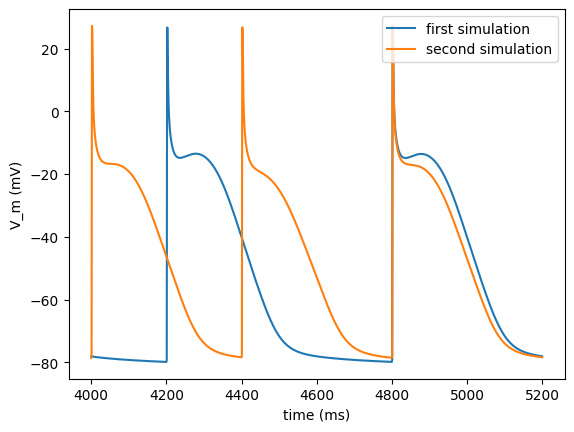
Basic functionality II
The second tutorial should familiarize you with futher basic functionality of the ionic cell model program bench.
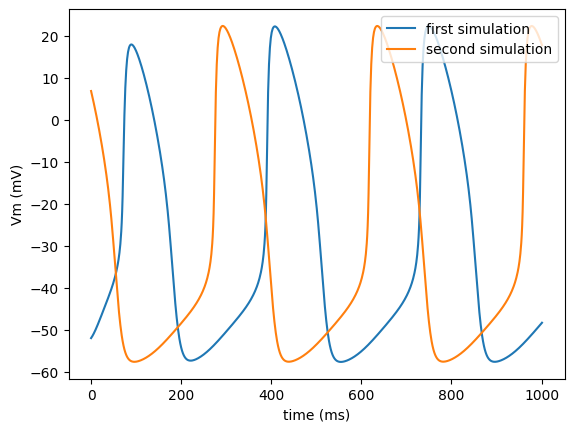
Advanced functionality I
The third single cell tutorial should familiarize you with some advanced functionality of the ionic cell model program bench.
Tissue Tutorials
Aim of the tissue tutorials is to make you familiar with the command line interface of the cardiac electrophysiology simuation program openCARP to develop tissue experiments. You will also learn further functionality of carputils. We strongy suggest to go through them in sequence.
Overview page of onboarding tutorials
If you want to start from the base and see this overview directly in openCARP JupyterLab.
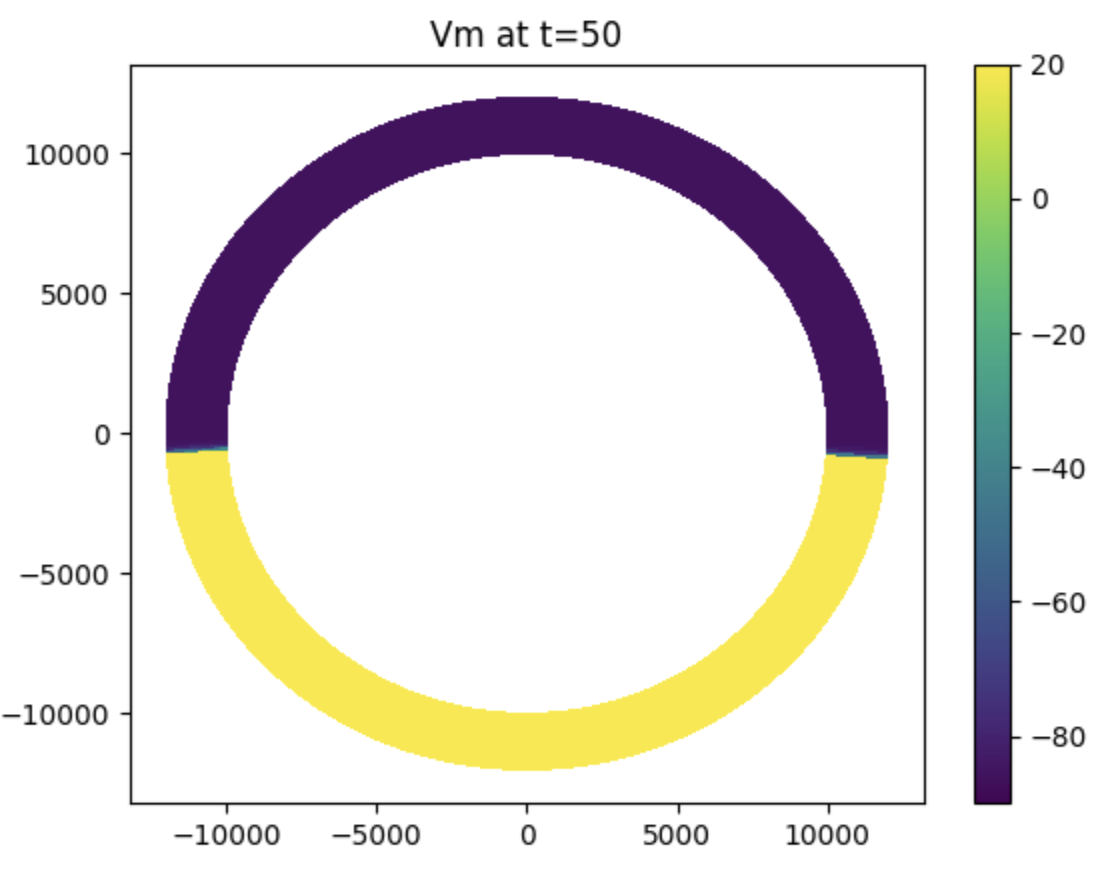
Basic tissue I
Available commands and models, running simple model and visualize results.
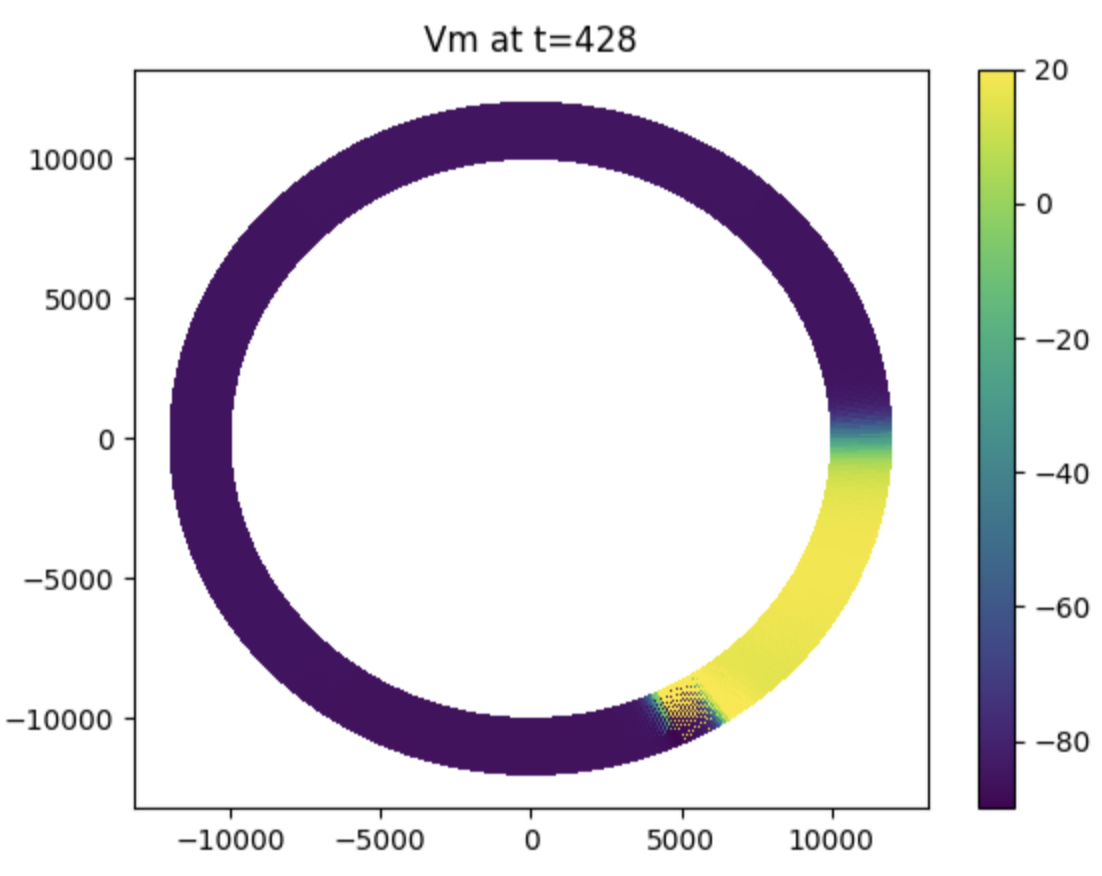
Basic tissue II
Adding another stimulus to generate a rotating wave in the ring model.

Basic tissue III
Applying carputils-style execution and adjust model parameters to induce re-entry.
Eikonal Tutorials
The aim of this tutorial is to teach the required parameters for running Eikonal/Reaction-Eikonal/DREAM simulations in openCARP.
Overview page of onboarding tutorials
If you want to start from the base and see this overview directly in openCARP JupyterLab.
Basic DREAM usage
Available commands and models, running a DREAM simulation with a simple pacing protocol.
