Tuning velocities and anisotropy ratios
See code in GitLab.
Author: Caroline Mendonca Costa caroline.mendonca-costa@kcl.ac.uk
To run the experiments of this tutorial do
cd ${TUTORIALS}/02_EP_tissue/03A__study_prep_tuneCVConcept
This tutorial introduces the theoretical background for the relationship between conduction velocity and tissue conductivity and how an iterative method to tune the conductivity values to yield a desired velocity for a given setup can be derived. In addition, the definition of anisotropy ratio and its impact on simulation results are presented. A simple strategy to enforce desired anisotropy ratio and conduction velocities is also presented.
Tuning conduction velocity
The relationship between conduction velocity and conductivity
A minimum requirement in modeling studies which aim at making case-specific predictions on ventricular electrophysiology is that activation sequences are carefully matched. Conduction velocity in the ventricles is orthotropic and may vary in space, thus profoundly influencing propagation patterns.
As described in Section [Tissue scale], the monodomain model [1] is equivalent to the bidomain model [2] if the monodomain conductivity tensor is given by the harmonic mean tensor, \({\sigma_m}\)
\[\mathbf{\sigma_m} = (\mathbf{\sigma_i}+\mathbf{\sigma_e})*(\mathbf{\sigma_i}*\mathbf{\sigma_e})^{-1},\]
\(\mathbf{\sigma_i}\) and \(\mathbf{\sigma_i}\) denoting the intra- and extracellular conductivity, respectively. Conduction velocity, CV, is not a parameter in the bidomain equations [2] and as such cannot be directly parametrized. However, assuming a continuously propagating planar wavefront along a given direction, \(\zeta\), one can derive a proportionality relationship 1 between \(CV_\zeta\) and \(\sigma_{m\zeta}\)
\[CV_\zeta \propto \sqrt{{\sigma_{m\zeta}}}\]
- For simplicity, the surface-to-volume ratio, \(eta\), was ommited in this tutorial,
-
as it is kept constant when tuning conductivities. 2
Conduction velocity as a function of simulation parameters
Experimental measurements of conductivity values are scarce and the variation in measured values across studies is vast. These uncertainties inevitably arise due to the significant degree of biological variation and the substantial errors in the measurement techniques themselves. Modeling and technical uncertainties may also have an impact on model predictions. Particularly, CV depends on the specific model used to describe cellular dynamics, \(I_\mathrm{ion}\), the spatial discretization, \(\Delta_x\), and on several implementation choices including the time-stepping scheme used, which we represent as \(\xi\). Thus, CV can be represented as a function
\[CV_\zeta = CV_\zeta({\sigma_{m\zeta}},I_\mathrm{ion},\Delta_x,\xi)\]
Iterative scheme for conduction velocity tuning
In most practical scenarios, \(I_\mathrm{ion}\), \(\Delta_x\), and \(\xi\) are parameters defined by users in the course of selecting a simulation software, an ionic model and a provided mesh to describe the geometry. Thus, only \({\sigma_{m\zeta}}\) is left which can be tuned to achieve a close match between the pre-specified conduction velocity, \(CV_\zeta\), and the velocity, \(\overline{CV_\zeta}\), predicted by the simulation.
Using equ-sigma-cv, and equ-sigma-m, one can find unique monodomain conductivities along all
axes \(\zeta\), which yield the prescribed conduction velocities, \(CV_\zeta\),
by iteratively refining conductivities based on \(\overline{CV_\zeta}\),
measured in simple 1D cable simulations. The iterative update scheme we propose is given as
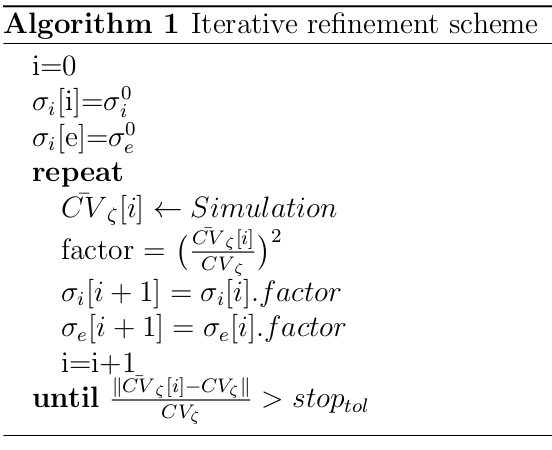
The algorithm is implemented as follows. Given an initial conductivity "guess", \({\sigma_i}^0\) and \({\sigma_e}^0\) and all other simulation parameters defined by \(I_\mathrm{ion}\), \(\Delta_x\), and \(\xi\), a simulation is run using a 1D cable and the conduction velocity is computed, as described in the figure below:
Using \(factor = (CV_{\zeta}/\overline{CV_{\zeta}})^2\), the conductivities \({\sigma_i}[i+1]\) and \({\sigma_e}[i+1]\) are updated. A new simulation is then run with the new conductivities. These steps are repeated until the error in CV is below a given tolerance, \(stop_\mathrm{tol}\).
Problem Setup
This tutorial will run a simulation on a 1D cable and compute the simulated CV, \(\bar{CV}\), and/or run
the iterative scheme shown Figure fig-algorithm to compute the conductivity \({\sigma_m}\) which yields
the prescribed CV for the chosen simulation setup.
Usage
The experiment specific options are:
--resolution RESOLUTION
Spatial resolution
--velocity VELOCITY Desired conduction velocity (m/s)
--gi GI Intracellular conductivity (S/m)
--ge GE Extracellular conductivity (S/m)
--ts TS Choose time stepping method. 0: explicit Euler, 1:
crank nicolson, 2: second-order time stepping
--dt DT Integration time step on micro-seconds
--converge {0,1} 0: Measure velocity with given setup or 1: Compute
conductivities that yield desired velocity with given
setup
--compareTuning run tuneCV with --resolution 100 200 and 400 with and
without tuning and make comparison plot
--compareTimeStepping
run tuneCV with --resolution 100 200 and 400 with
Explicit Euler and Crank Nicolson and make comparison
plot
--compareMassLumping run tuneCV with --resolution 100 200 and 400 with and
without mass lumping and make comparison plot
--compareModelPar run tuneCV with --resolution 100 200 and 400 with
original Ten Tusher cell model and with reduced
sodium conductance and make comparison plot
--ar AR run tuneCV with for CV_f = 0.6 and CV_s = 0.3 m/s with
given anisotropy ratio (ar)The user can either run single experiments, do a comparison of the multiple
resolutions with the options --compareTuning, --compareTimeStepping, --compareMassLumping, and --compareModelPar,
or compare the change in conductivities with different anisotropy ratios.
HINT Since all --compare options try to write into an existing directory at some point, the runs might seem to stop and get stuck in the terminal. Simply press ENTER once to continue to the overwrite check, type y and ENTER again to continue the simulations.
Run 1D examples
To run the experiments of this tutorial do
cd ${TUTORIALS}/02_EP_tissue/03A__study_prep_tuneCVcompareTuning:
To compute the conductivities for resolutions of 100, 200, and 400 um with and without tuning, run: For webGUI, remove RESOLUTION value
./run.py --compareTuning --visualizeAfter running this command you should see the following plot
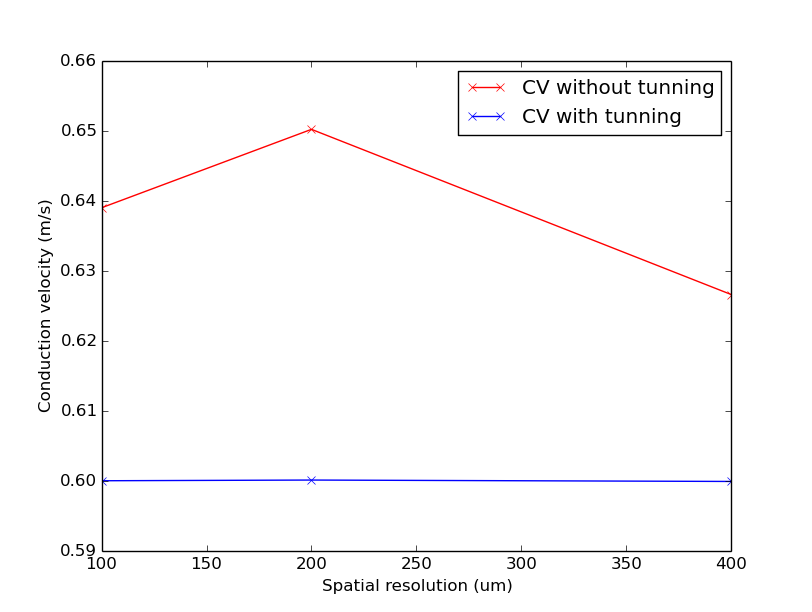
Notice that CV varies with resolution in a non-linear fashion and CV is constant when the iterative scheme is applied.
compareTimeStepping:
To compare the effect using the explicit Euler or Crank–Nicolson methods for the resolutions of 100, 200, and 400 um run: For webGUI, remove RESOLUTION and TS values
./run.py --compareTimeStepping --visualize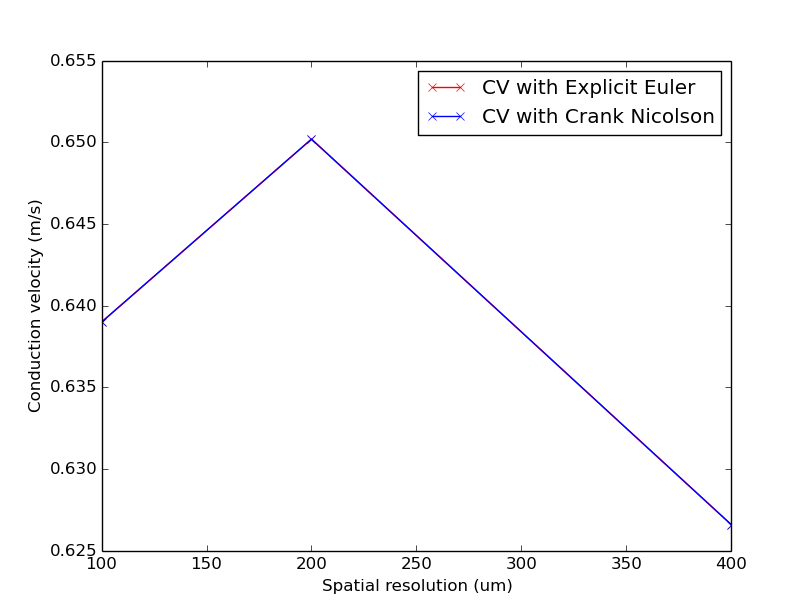
Notice that CV varies with resolution virtually identically with both time-stepping schemes.
compareMassLumping:
Mass lumping is a common numerical technique in FEM to speed up computation. The mass matrix, \(M\), is made diagonal, implying that its inverse is also diagonal, and solving the system is trivial. The lumped mass matrix, \(M^L\), is computed by setting its main diagonal to be
\[M_{ii}^L = \Sigma_{j=1}^N M_{ij}\]
To compare the effect of using mass lumping on CV for the resolutions of 100, 200, and 400 um run: For webGUI, remove RESOLUTION and LUMPING values
./run.py --compareMassLumping --visualize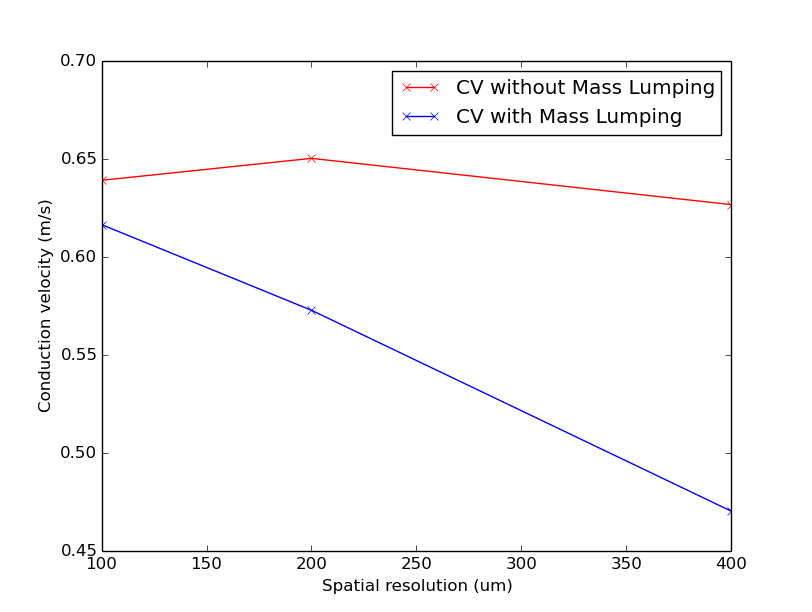
Notice that CV varies with resolution in both cases, but the decrease in CV for coarser resolutions is much steeper with mass lumping.
compareModelPar:
To compare the effect of modifying the sodium conduction on CV for the resolutions of 100, 200, and 400 um run: For webGUI, remove RESOLUTION, MODEL and MODELPAR values
./run.py --compareModelPar --visualize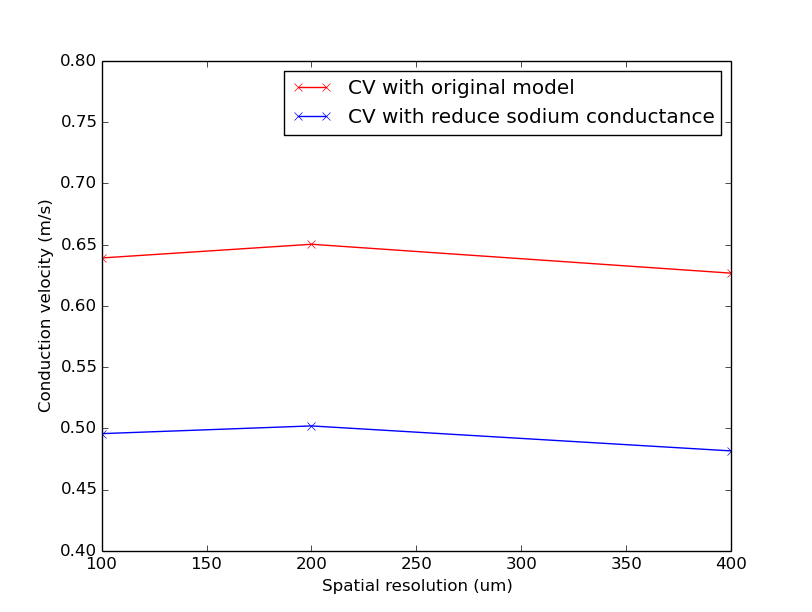
Notice that CV varies with resolution similarly in both cases, but the CV with reduced sodium conductance is lower than with the original model.
Run 2D and 3D examples
As previously mentioned, conduction velocity in the ventricles is orthotropic, that is, CV is different in each axis \(\zeta\). Estimated average CVs in the longitudinal, transverse, and sheet normal directions, \(CV_f\), \(CV_s\) and \(CV_n\), are about 0.67 m/s, 0.3 m/s, and 0.17 m/s, respectively. 4
2D:
To run a 2D simulation where \(CV_f > CV_s\), that is, an anisotropic setup, two sets of conductivities must be defined, one for each axis \(\zeta=f,s\). To compute two sets of conductivities with tuneCV, call the example without the "compare" flags twice with two different velocities and look at the conductivities output on the screen.
For the longitudinal direction, run
./run.py --velocity 0.6 --converge 1You should get an output close to the following:
Conduction velocity: 0.6398 m/s [gi=0.1740, ge=0.6250, gm=0.1361]
Conduction velocity: 0.6006 m/s [gi=0.1530, ge=0.5497, gm=0.1197]
Conduction velocity: 0.6001 m/s [gi=0.1527, ge=0.5486, gm=0.1195]where gi, ge, and gm are \({\sigma_i}\), \({\sigma_e}\), and \({\sigma_m}\), respectively. The \({\sigma_i}\) and \({\sigma_e}\) pair will yield a CV of 0.6 m/s in both monodomain and bidomain simulations.
Now, for the transverse direction, run
./run.py --velocity 0.3 --converge 1You should get an output close to the following:
Conduction velocity: 0.6398 m/s [gi=0.1740, ge=0.6250, gm=0.1361]
Conduction velocity: 0.3070 m/s [gi=0.0383, ge=0.1374, gm=0.0299]
Conduction velocity: 0.3001 m/s [gi=0.0365, ge=0.1312, gm=0.0286]
Conduction velocity: 0.3000 m/s [gi=0.0365, ge=0.1311, gm=0.0285]In this case, the \({\sigma_i}\) and \({\sigma_e}\) pair will yield a CV of 0.3 m/s.
3D:
Similarly, to run a 3D simulation where \(CV_f > CV_s > CV_n\), that is, an orthotropic setup, three sets of conductivities must be defined, one for each axis \(\zeta=f,s,n\). To compute the set of conductivities for the sheet normal direction, \(\zeta=n\), call the example once more with \(CV_n = 0.2\)
./run.py --velocity 0.2 --converge 1You should get an output close to the following:
Conduction velocity: 0.6398 m/s [gi=0.1740, ge=0.6250, gm=0.1361]
Conduction velocity: 0.2038 m/s [gi=0.0170, ge=0.0611, gm=0.0133]
Conduction velocity: 0.1998 m/s [gi=0.0164, ge=0.0588, gm=0.0128]
Conduction velocity: 0.2000 m/s [gi=0.0164, ge=0.0590, gm=0.0128]In this case, the \({\sigma_i}\) and \({\sigma_e}\) pair will yield a CV of 0.2 m/s.
- Notice that for \(CV_s = 0.3\) and \(CV_n = 0.2\), four iterations were required to converge to
-
the desired CV, whereas \(CV_f = 0.6\) required three. This is because, in this example, the initial conductivity \({\sigma_{m,\zeta}}^0\) (see Figure
fig-algorithm) yields a velocity close to 0.6 m/s and is the same in all cases. Thus, the initial error in CV is larger for \(CV_s=0.3\) and \(CV_n=0.2\) than for \(CV_f=0.6\).
Tuning Anisotropy Ratios
Defining anisotropy ratios
According to experimental measurements, the intracellular domain is more anisotropic than the extracellular domain, that is, the ratio between the longitudinal and transverse conductivities is larger in the intracellular space than in the extracellular space.
In the intracellular domain, the anisotropy ratio between the longitudinal, \(\zeta=f\), and transverse, \(\zeta=s\), directions is defined as
\[\alpha_{ifs} = {\sigma_{if}}/{\sigma_{is}}\]
where \({\sigma_{if}}\) and \({\sigma_{is}}\) are the intracellular conductivities in the longitudinal and transverse direction, respectively.
Similarly, in the extracellular domain, the anisotropy ratio between the longitudinal, \(\zeta=f\), and transverse, \(\zeta=s\), directions is defined as
\[\alpha_{efs} = {\sigma_{ef}}/{\sigma_{es}}\]
where \({\sigma_{ef}}\) and \({\sigma_{es}}\) are the extracellular conductivities in the longitudinal and transverse direction, respectively.
Now, we can express the anisotropy ratio between the two domains as in the longitudinal and transverse directions as
\[\alpha_{lt} = \alpha_{ifs}/\alpha_{efs} = ({\sigma_{if}}*{\sigma_{es}})/({\sigma_{ef}}*{\sigma_{is}})\]
Computing conductivities with a fixed anisotropy ratio
There are many ways to enforce anisotropy ratios between conductivity values. In this tutorial, we use a
fixed anisotropy strategy, where, given an anisotropy ratio, we compute an initial transverse conductivity
for extracellular domain. The intracellular conductivities as well as the longitudinal extracellular
conductivity are kept fixed at default values. Using Equation equ-aniso we obtain:
\[{\sigma_{es}} = (\alpha_{lt}*{\sigma_{ef}}*{\sigma_{is}})/{\sigma_{if}}\]
We then tune the longitudinal and transverse conductivities separately using the iterative scheme. In this manner, we enforce both the desired CV and anisotropy ratio.
The effect of equal and unequal anisotropy ratios
If \(\alpha_{ifs} = \alpha_{efs}\), then \(\alpha_{lt} = 1\). In this case, the two domains have equal anisotropy ratios. On the other hand, if \(\alpha_{ifs} > \alpha_{efs}\), then \(\alpha_{lt} > 1\). In this case, the two domains have unequal anisotropy ratios. Unequal anisotropy ratios can only be represented with the bidomain model [2].
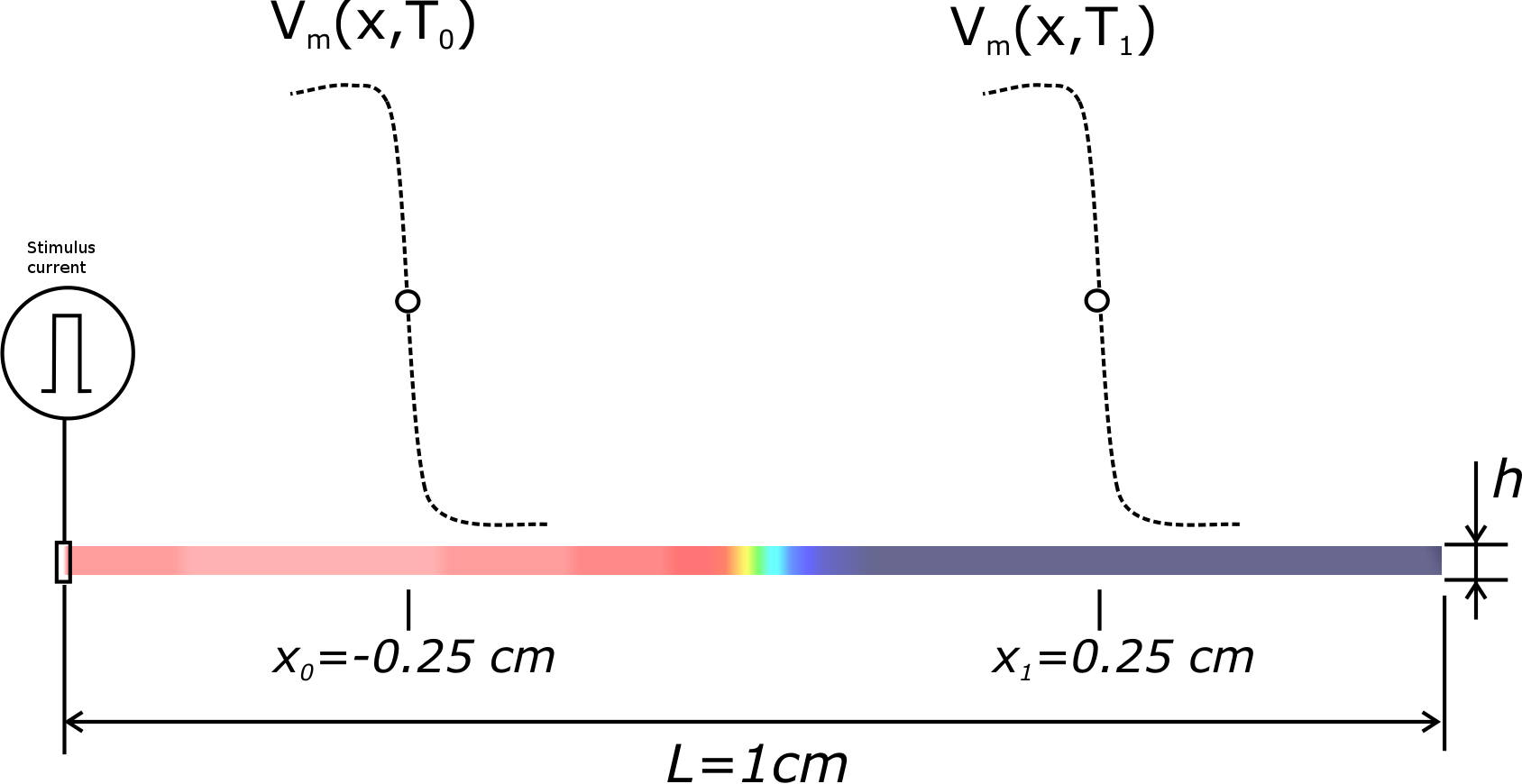
While anisotropy ratios are of rather minor relevance when simulating impulse propagation in tissue 5 , they play a prominent role when the stimulation of cardiac tissue via externally applied electric fields is studied. In this case, virtual electrodes appear around the stimulus site, as show in the figure below.
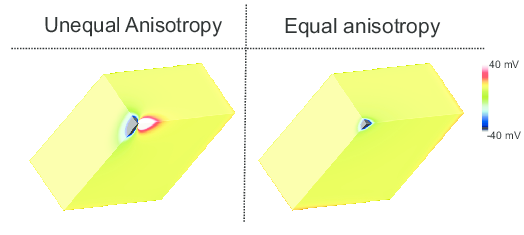
Run anisotropy ratio examples
To compute longitudinal and transverse conductivities for the case of equal anisotropy ratios and \(CV_f = 0.6\) and \(CV_s = 0.3\), run For webGUI, remove VELOCITY, GI, GE and CONVERGE values
./run.py --ar=1.0You should get conductivities similar to these:
g_if: 0.152700 g_is: 0.036500 g_ef: 0.548500 g_es: 0.131100To compute longitudinal and transverse conductivities for the case of unequal anisotropy ratios and \(CV_f = 0.6\) and \(CV_s = 0.3\), run
./run.py --ar=3.0You should get conductivities similar to these:
g_if: 0.152700 g_is: 0.031200 g_ef: 0.548500 g_es: 0.336100References
Costa CM, Hoetzl E, Rocha BM, Prassl AJ, Plank G., Automatic Parameterization Strategy for Cardiac Electrophysiology Simulations, Comput Cardiol 40:373-376, 2013. [Pubmed]↩︎
Costa CM, Hoetzl E, Rocha BM, Prassl AJ, Plank G., Automatic Parameterization Strategy for Cardiac Electrophysiology Simulations, Comput Cardiol 40:373-376, 2013. [Pubmed]↩︎
Costa CM, Hoetzl E, Rocha BM, Prassl AJ, Plank G., Automatic Parameterization Strategy for Cardiac Electrophysiology Simulations, Comput Cardiol 40:373-376, 2013. [Pubmed]↩︎
B. J. Caldwell, M. L. Trew, G. B. Sands, D. A. Hooks, I. J. LeGrice, B. H. Smaill, Three distinct directions of intramural activation reveal nonuniform side-to-side electrical coupling of ventricular myocytes., Circ Arrhythm Electrophysiol, vol. 2, no. 4, pp. 433-440, Aug 2009. [Pubmed]↩︎
Costa CM, Hoetzl E, Rocha BM, Prassl AJ, Plank G., Automatic Parameterization Strategy for Cardiac Electrophysiology Simulations, Comput Cardiol 40:373-376, 2013. [Pubmed]↩︎
Recent questions tagged experiments, examples, tunecv, conductivity, tissue
There are tagged with experiments, examples, tunecv, conductivity, tissue.
Here we display the 5 most recent questions. You can click on each tag to show all questions for this tag.
You can also ask a new question.
