Electrical Mapping
See code in GitLab.
Author: Fernando Campos fernando.campos@kcl.ac.uk
This example demonstrates how to compute local activation times (LATs) and action potential durations (APDs) of cells in a cardiac tissue. To run the experiments of this example change directories as follows:
cd ${TUTORIALS}/02_EP_tissue/08_latsElectrical Mapping
The wavelength of the cardiac impulse, given by the product of conduction velocity (CV) and refractory period, is of utmost importance for the study of arrhythmia mechanisms. The assessment of the CV as well as the refractory period of the cardiac action potential (AP) relies on the determination of activation and repolarization times, respectively. Experimentally, these are usually calculated based on 1) transmembrane voltages \(V_{\mathrm m}\) measured by glass micro-electrodes or in optical mapping; or 2) extracellular potentials \(\phi_{\mathrm e}\) measured at the surface of the tissue.
The estimation of activation/repolarization times is based on an event detector that records the instants when the selected event type occurs. Local activation time (LAT), for instance, is usually taken as the time of maximum \(\mathrm{d}V_{\mathrm m}/\mathrm{d}t\) or the minimum \(\mathrm{d}\phi_{\mathrm e}/\mathrm{d}t\):
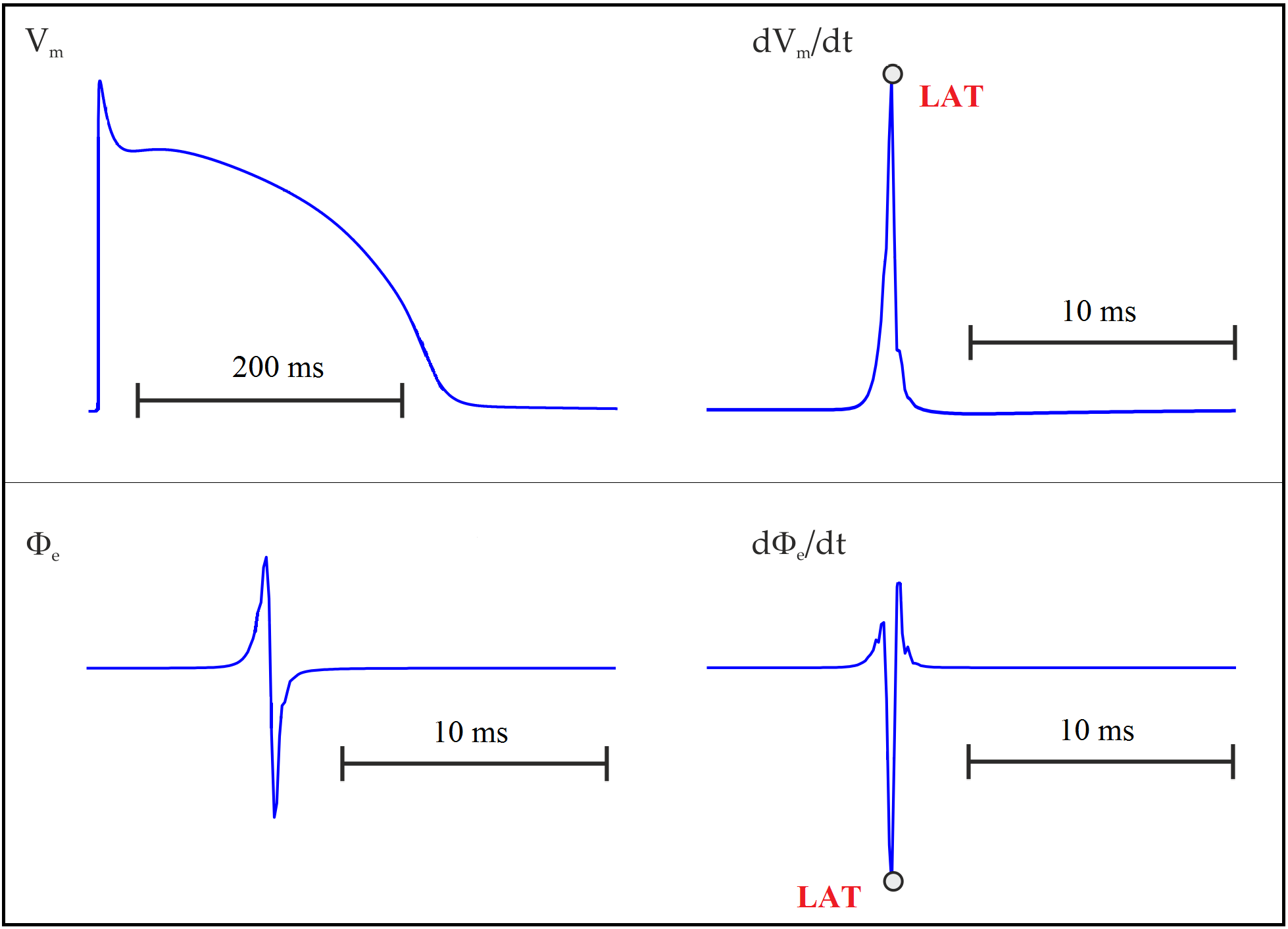
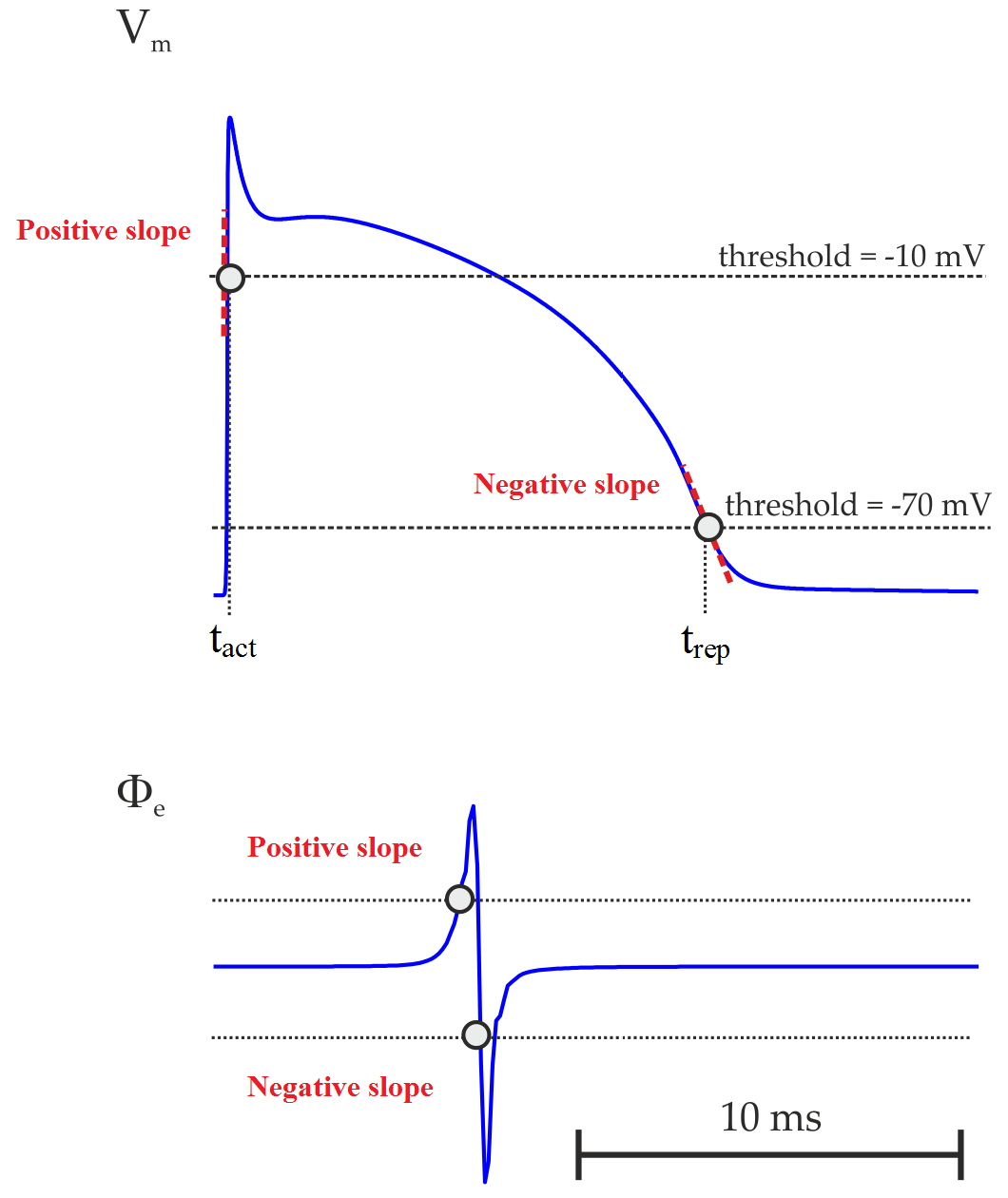
Instead of using derivatives, LATs can also been estimated by the crossing of a threshold value provided that the chosen value reflects the sodium channel activation since they are responsible for the rising phase of AP. One could, for instance, pick the time instant when \(V_{\mathrm m}\) crosses -10 mV with a positive slope. Equally, one could look for crossing of \(V_{\mathrm m}\) with a negative slope to detect repolarization events. AP duration (APD) can then be computed as the difference between repolarization and activation times. The detection of activation (\(t_{\mathrm{act}}\)) and repolarization (\(t_{\mathrm{rep}}\)) times based on threshold crossing is illustrated below:
Problem Setup
This example will run a simulation using a 2D sheet model (1 cm x 1 cm). A transmembrane stimulus current of 100 uA/cm^2 is applied for a period of 1 ms at the middle of the left side of the tissue. A ground electrode is placed at the opposite side of the stimulating electrode. Simulation of electrical activity is based on the bidomain equations. Dynamics of the cardiac myocytes within the tissue are described using the Modified Beeler-Router Drouhard-Roberge (DrouhardRoberge) model. Activation times are obtained either from \(V_{\mathrm m}\) or \(\phi_{\mathrm e}\) using a threshold crossing method, whereas repolarization can only be obtained by threshold crossing from \(V_{\mathrm m}\). Estimation of repolarization times from \(\phi_{\mathrm e}\) is out of the scope of this example.
Usage
To run this example, execute:
cd ${TUTORIALS}/02_EP_tissue/08_lats
./run.py --help The following optional arguments are available (default values are indicated):
--quantity Quantity/variable/signal used to compute activation time
Options: {vm,phie}
Default: vm
--act-threshold The threshold value for determining activation from vm or phie
Default: -10 mV
--rep-threshold The threshold value for determining repolarization from vm
Default: -70 mV
--show-APDs Visualize APDs instead of LATsIf the program is run with the --visualize option, meshalyzer will automatically load the computed
activation sequence (LATs). If the --show_APDs is given, then the computed APDs will be loaded:
Visualize LATs:
./run.py --visualize
Visualize APDs:
./run.py --show-APDsInterpreting Results
Differences between activation sequences computed from \(V_{\mathrm m}\) or \(\phi_{\mathrm e}\)
and/or with different thresholds can be appreciated with meshalyzer (--visualize). The activation
sequence for the default parameters looks like this:
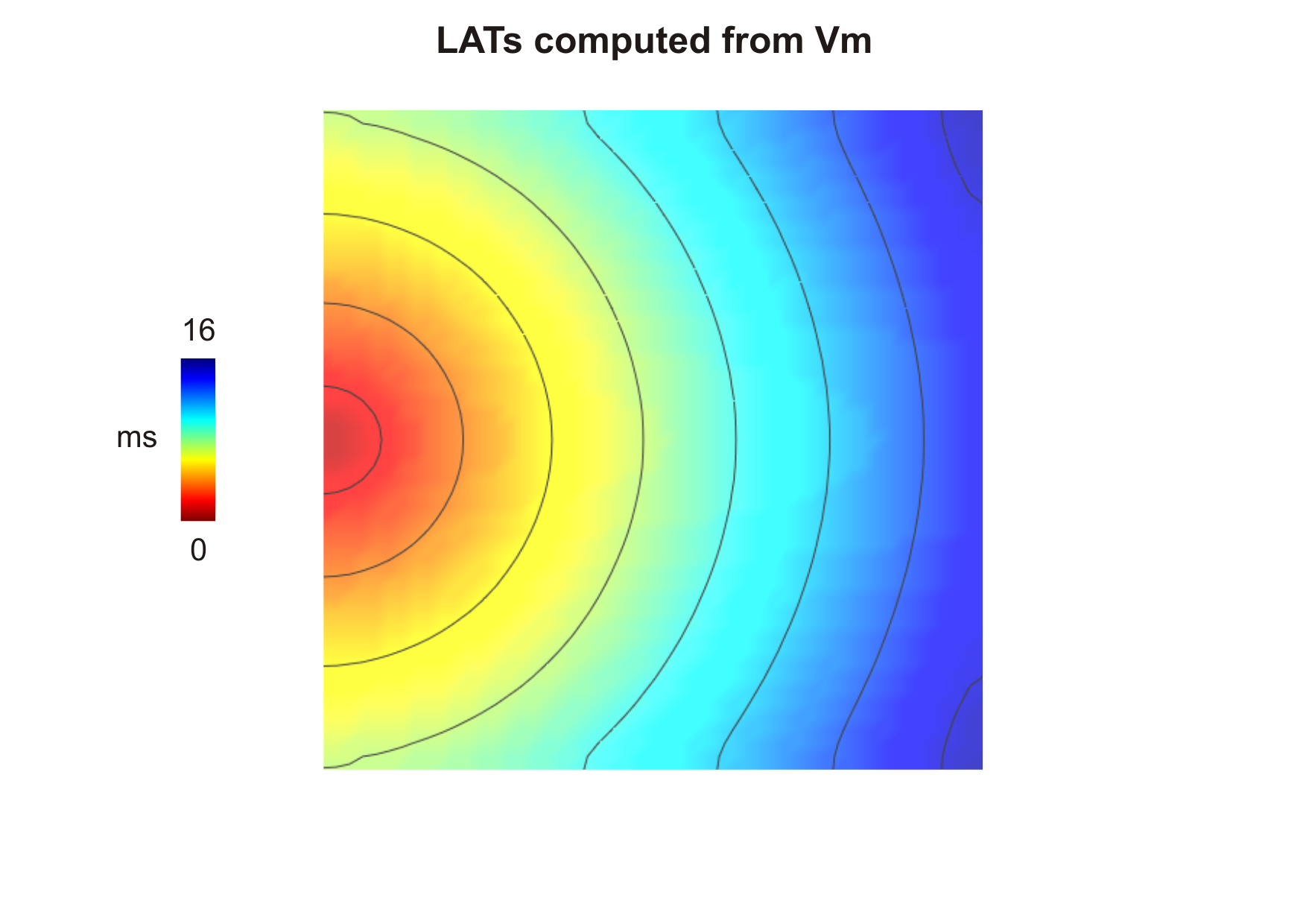
--act-threshold=-10) obtained after
delivering a stimulus current in the middle of the left side tissue.Note
- It is important to note that while computing activation from \(V_{\mathrm m}\) -lats[].mode has to be set to zero. This is because \(V_{\mathrm m}\) is about -85 mV in cardiac cells at rest and rapidly rises to about +40 mV during the activation phase. This means \(V_{\mathrm m}\) will cross the threshold with a positive slope. While computing repolarization, on the other hand, \(V_{\mathrm m}\) goes from positive values back to the rest. The threshold will be then crossed with a negative slope.
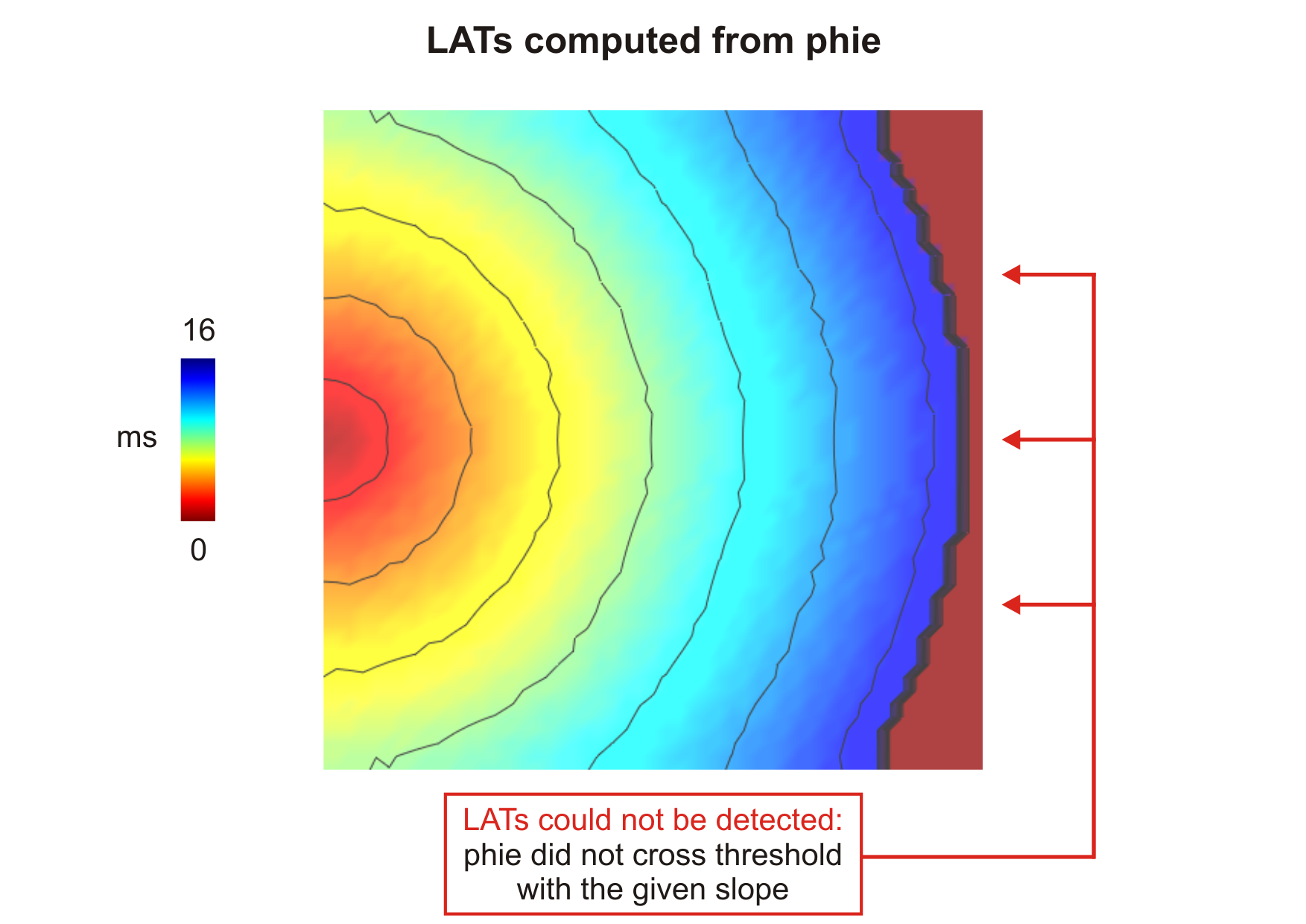
--act-threshold=-10) obtained
after delivering a stimulus current in the middle of the left side tissue.Note
- Unlike the shape of \(V_{\mathrm m}\) which remains pretty much the same in all cells, \(\phi_{\mathrm e}\) morphology may vary depending on its position in relation to both stimulation and ground electrodes. Thus, LATs obtained based on \(\phi_{\mathrm e}\) might differ from those obtained from \(V_{\mathrm m}\).
- If the criteria for threshold crossing detection is never satisfied, the resulting entry in the output file will be -1. See action sequence obtained from \(\phi_{\mathrm e}\).
The computed APDs using the default parameters looks like this (use --show-APDs instead of --visualize):
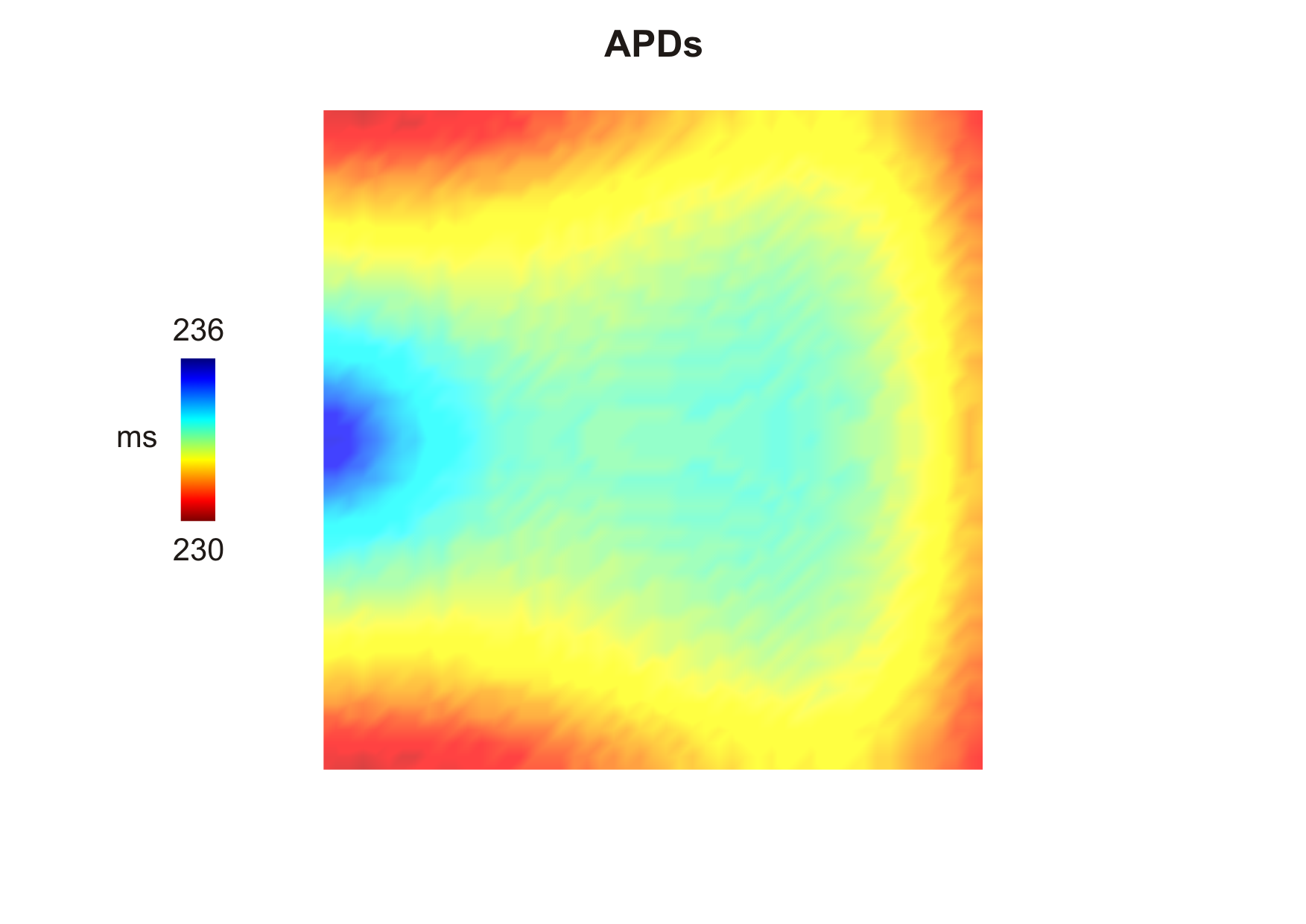
--act-threshold=-10 and
--rep-threshold=-70) after delivering a stimulus current in the middle of the left side of
the tissue.openCARP Parameters
The relevant parameters used in the tissue-scale simulation are shown below:
# Number of events to detect
-num_LATs = 2
# Event 1: activation
-lats[0].ID = 'ACTs'
-lats[0].all = 0
-lats[0].measurand = 0
-lats[0].threshold = -10
-lats[0].mode = 0
# Event 2: repolarization
-lats[1].ID = 'REPs'
-lats[1].all = 0
-lats[1].measurand = 0
-lats[1].threshold = -70
-lats[1].mode = 1num_LATs referes to the number of events we want to detect. In this example there are two events: activation and repolarization. -lats[0] are options to compute activation while -lats[1] are options to compute repolarization. -lats[].ID is the name of output file; -lats[].all tells openCARP if it should detect only the first event (-lats[].all = 0), i.e., first beat, of all of them (-lats[].all = 1); -lats[].measurand refers to the quantity: \(V_{\mathrm m}\) (0) or \(\phi_{\mathrm e}\) (1); -lats[].threshold is the threshold value for determining the event; finally, -lats[].mode tells openCARP to look for threshold crossing with a either a positive or negative slope.
Recent questions tagged experiments, examples, tissue, lat
There are tagged with experiments, examples, tissue, lat.
Here we display the 5 most recent questions. You can click on each tag to show all questions for this tag.
You can also ask a new question.
