Fitting the foot current
See code in GitLab.
Author: Cristian Barrios Espinosa cristian.espinosa@kit.edu
Finding Ifoot Parameters
Reaction eikonal simulations in openCARP are used to fit the parameters for the Ifoot current, which is essential to trigger the action potential [Neic2017]. The choice of parameters depends on the ionic model used, offering a computationally efficient way to calculate activation times from electrical wave propagation in cardiac tissue. These simulations do not model transmembrane voltage dynamics, making them ideal for fast approximations or pre-analysis steps.
To run the experiments described in this tutorial, navigate to the following directory:
cd ${TUTORIALS}/03_Eikonal/04_Fit_IfootSimulation Setup
To find the best fit for the Ifoot parameters, a monodomain simulation is first run using the desired ionic model and its parameters in a 3D block. The threshold voltage, V_th, must be provided. This threshold represents the minimum transmembrane voltage required to trigger an action potential, typically linked to the sodium channels' activation threshold, although more phenomenological models may adopt a simplified formulation.
python3 ./run.py --help
...
Tutorial specific options:
--imp IMP Ionic Model to optimize. (default: Courtemanche)
--model_par MODEL_PAR Ionic Model parameters. (default: )
--V_th V_TH Voltage threshold for the cutoff of the foot current (mV). (default: -30)Experiments
Experiment 1: Find the Ifoot parameters for the Courtemanche model (default)
The first experiment finds the Ifoot parameter for the Courtemanche model with default parameters [Courtemanche1998].
python3 run.pyThe output will provide the fitted parameters for the Ifoot model corresponding to the ionic model used, as well as the error of the fit:
Fitted A_F: 2.464218456421841
Fitted tau_F: 0.43917740479584627
RMSE: 0.687515024630087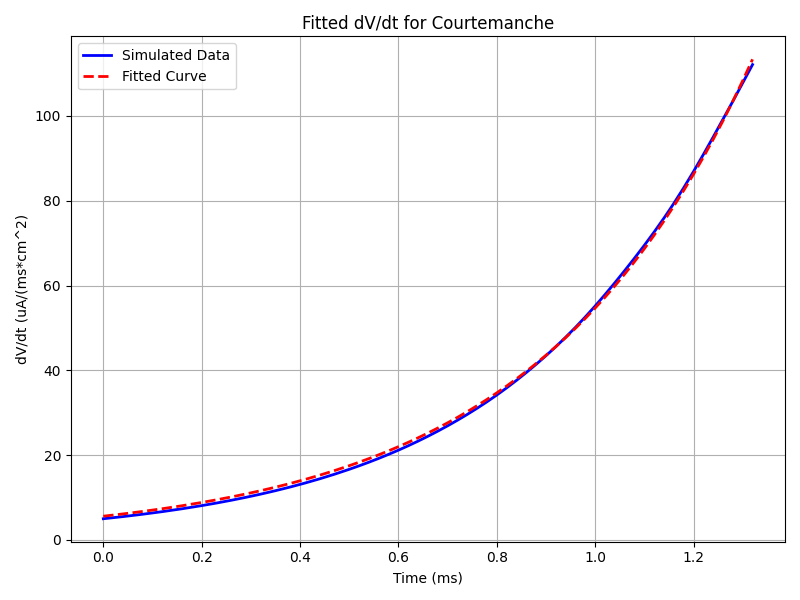
Experiment 2: Find the Ifoot parameters for the OHara-Rudy model
In this case, we change the ionic model and find the Ifoot parameters for the default set of parameters of [OHara2011].
python3 run.py --imp OHaraThe result should provide the fitted parameters as well:
Fitted A_F: 3.007586177286489
Fitted tau_F: 0.3141233370949477
RMSE: 2.3670844102567106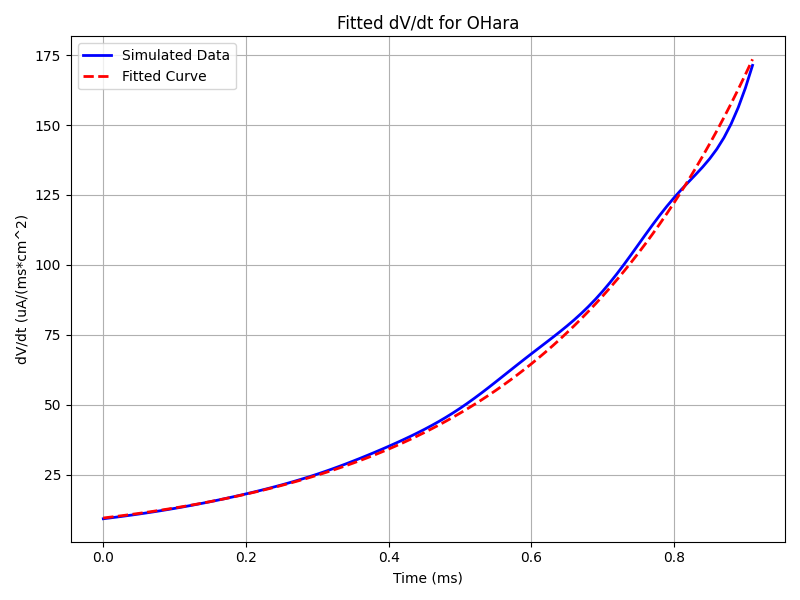
Experiment 3: Change model parameters
Modify the parameters of the Courtemanche model to simulate atrial fibrillation, as described by [Loewe2016].
run.py --model_par 'GCaL*0.45,GK1*2,GNa*1,factorGKur*0.5,Gto*0.35,GKs*2,maxIpCa*1.5,maxINaCa*1.6'The output should give the following result:
Fitted A_F: 4.025919988860185
Fitted tau_F: 0.4572368218676241
RMSE: 0.821926533960846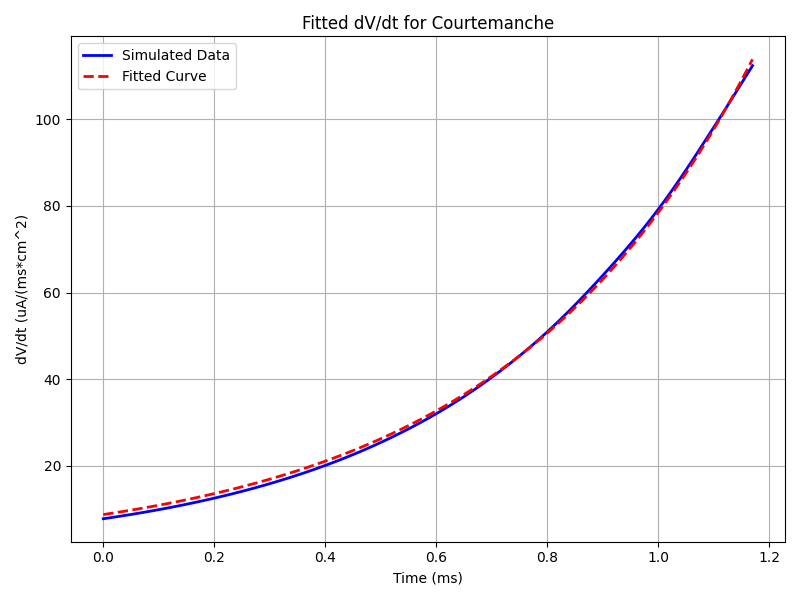
Recent questions tagged experiments, examples, tissue, eikonal, ifoot
There are tagged with experiments, examples, tissue, eikonal, ifoot.
Here we display the 5 most recent questions. You can click on each tag to show all questions for this tag.
You can also ask a new question.
